Software Development
IMAGE PLATFORM
- Home >
- Research >
- Technological platforms >
- Image >
- Software Development. Platform Image
- Image Analysis and Quantification Support Service
- Analysis Workstation (Room B37)
- Analysis Workstation (Room B02)
Responsible:
Tomás Muñoz: tmsantoro@unav.es
Phone: +34 948 194 700 Ext. 1043
Biomedical and medical image analysis
We develop software tools tailored to each user, adapted to each specific problem.
Most applications are developed as macros or plugins for Fiji/ImageJ. It is a free software platform, which allows the user to make use of our applications from any computer and perform their own image analysis and quantifications.
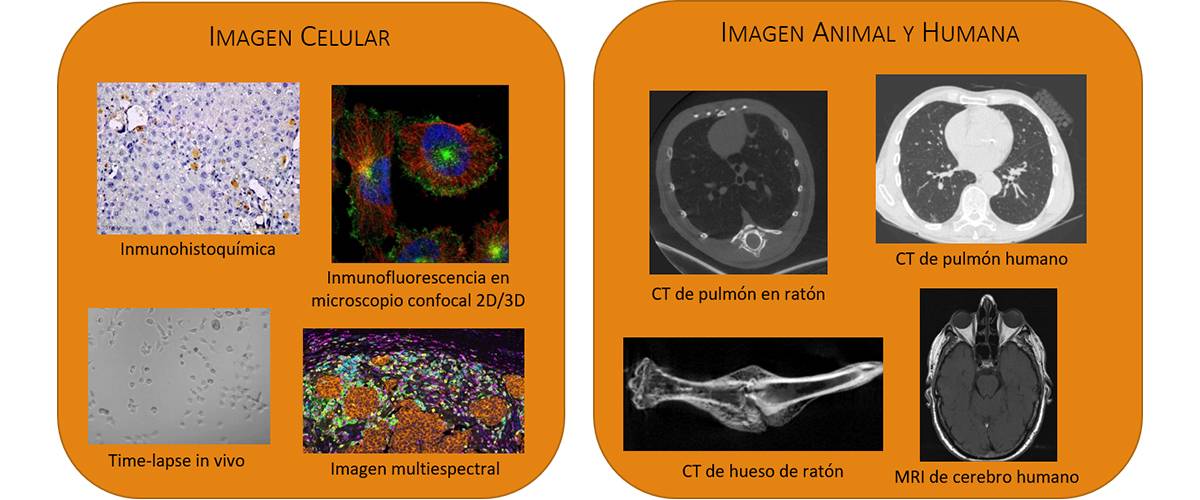
We work with different image modalities
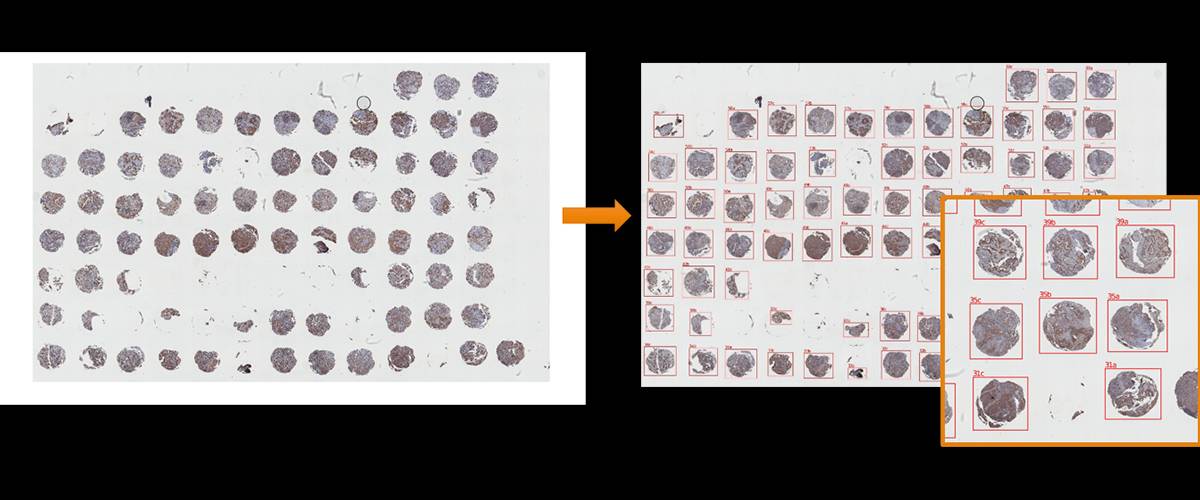
Automatic extraction of individual cores from full TMA images.
Individual cores are extracted as single images for further processing by other Fiji plugins.
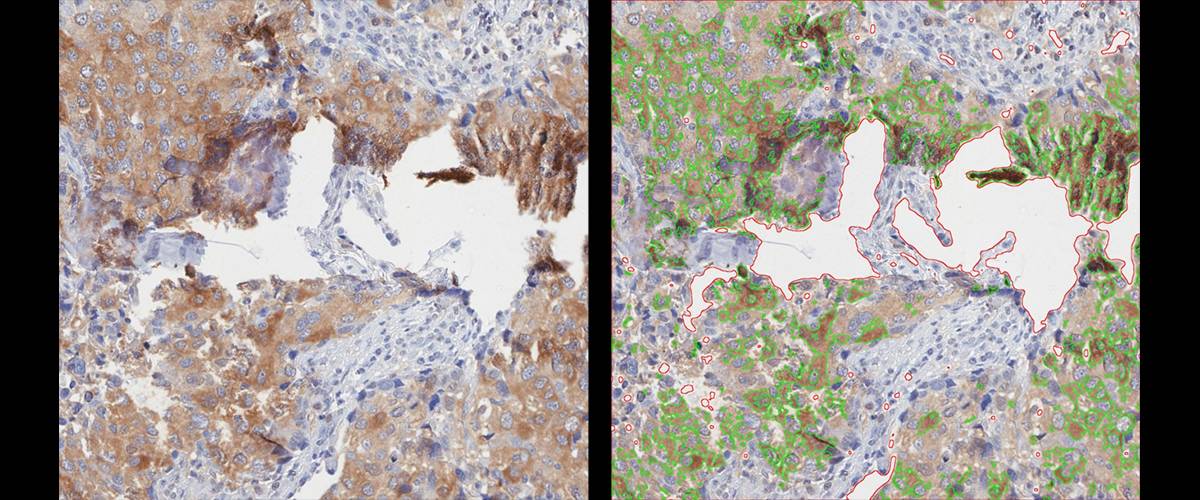
Detection and quantification of DAB immunostaining. Quantification of positive area to tissue area ratio. Possibility of manual removal of regions (necrotic areas, folds, etc.).
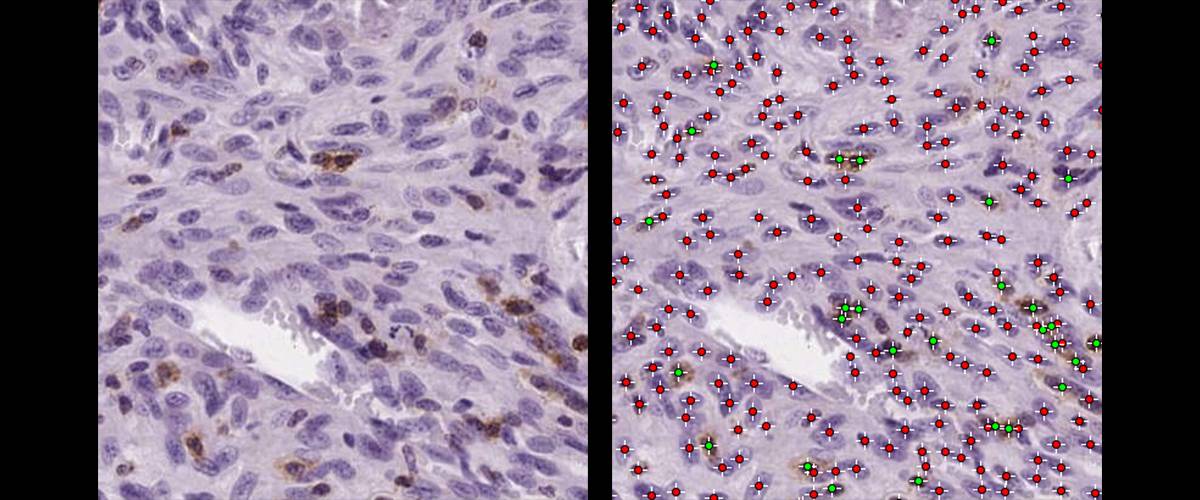
Detection and classification of single cells in H&E DAB images. Sorting and counting of DAB positive or negative cells.
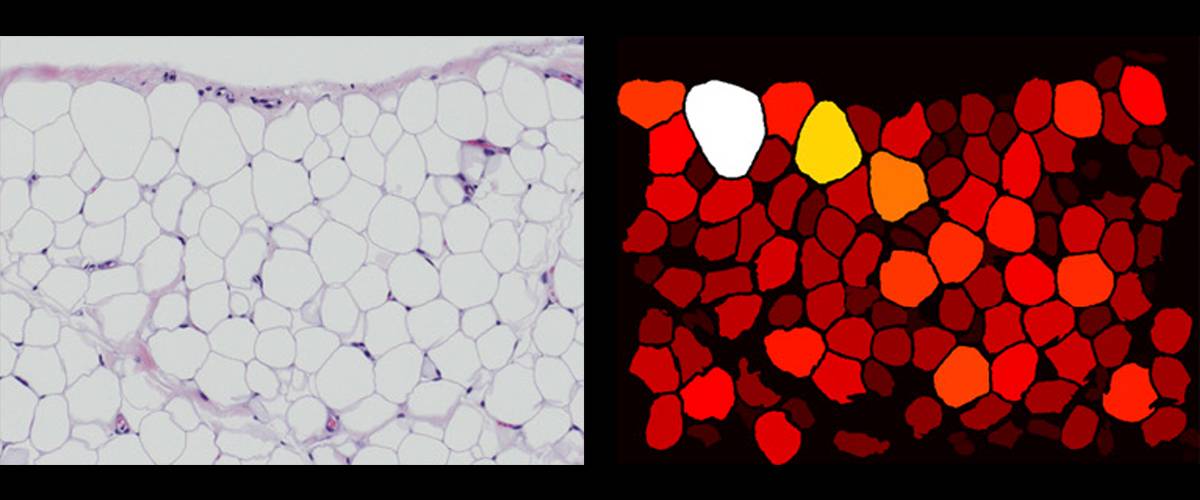
Adiposoft: automated software for the analysis of cellularity in white adipose tissue in histological sections. Segmentation of individual adipocytes. Morphological quantification of each adipocyte to obtain the complete distribution.
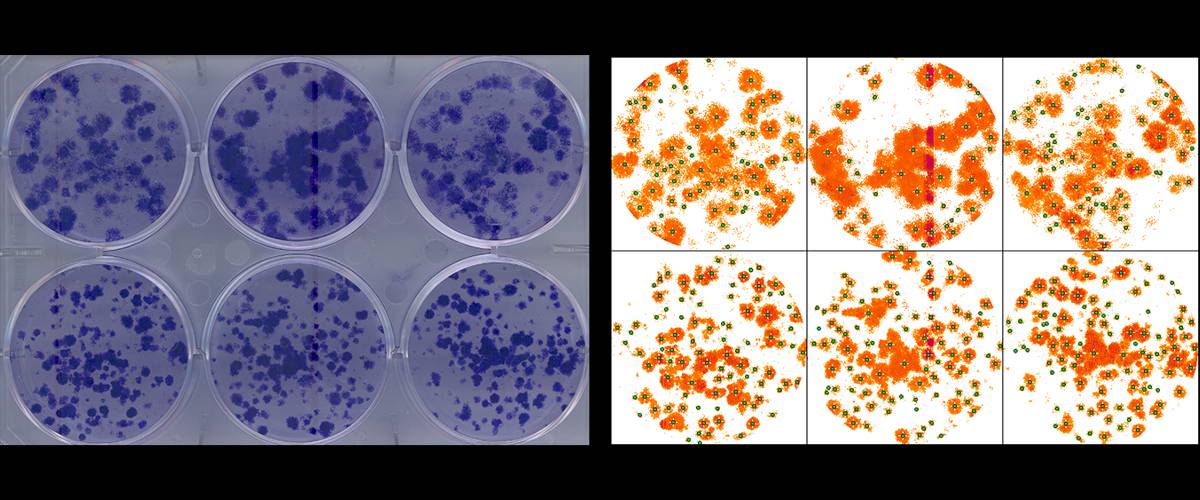
Analysis of colony formation assays in cell culture plates. Adaptation of the "ColonyArea" plugin by Guzman et al. 2014. Semi-automatic detection of individual wells, segmentation and quantification of area and number of colonies formed in each well.
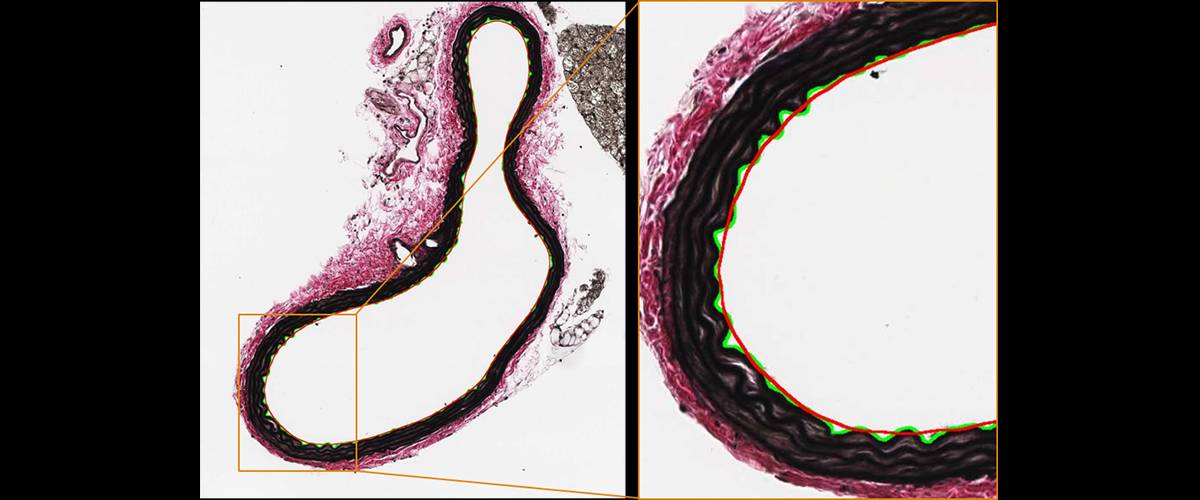
Analysis of elastin waviness in arteries. Automatic detection of the artery lumen, elastin ripples in the inner contour, approximation of a contour without ripples and calculation of the length ratio between both contours: rippled (green) vs. approximation without ripples (red).
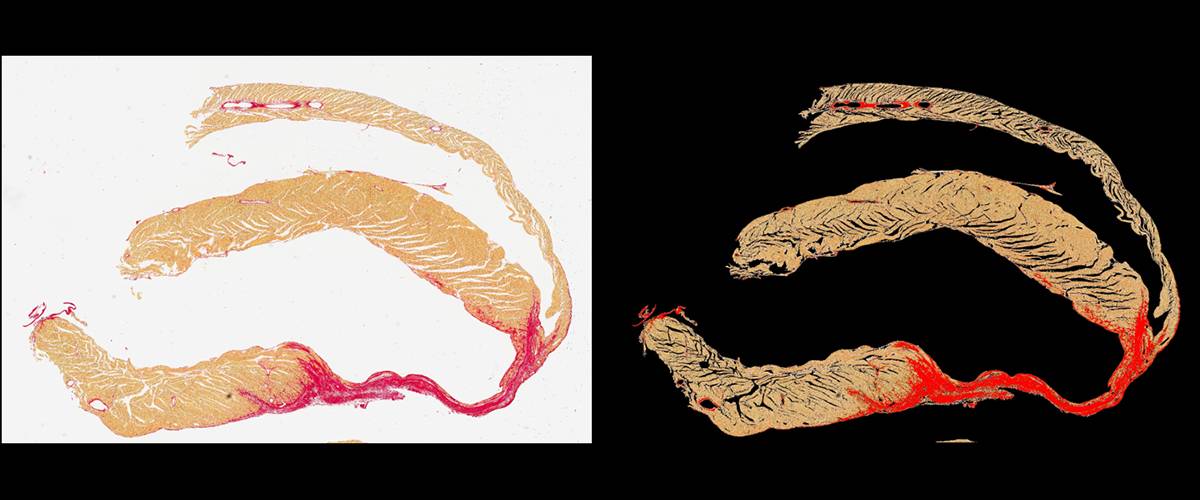
Quantification of fibrotic tissue by Sirius Red staining. Automatic detection of tissue and regions with positive Syrian Red staining. Calculation of ratio of positive area vs. total tissue area.
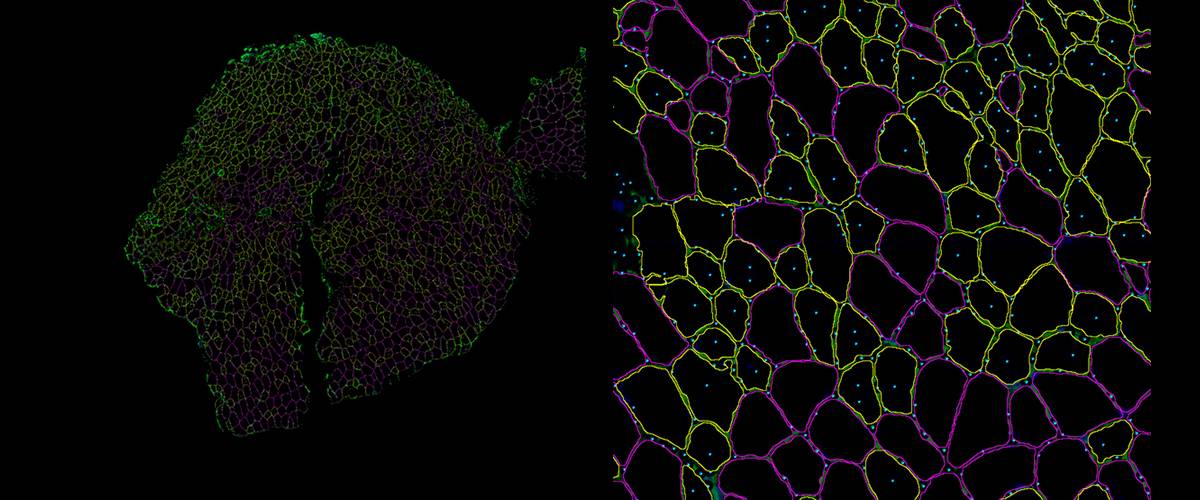
Automatic detection, classification and quantification of muscle fibers in immunofluorescence images.
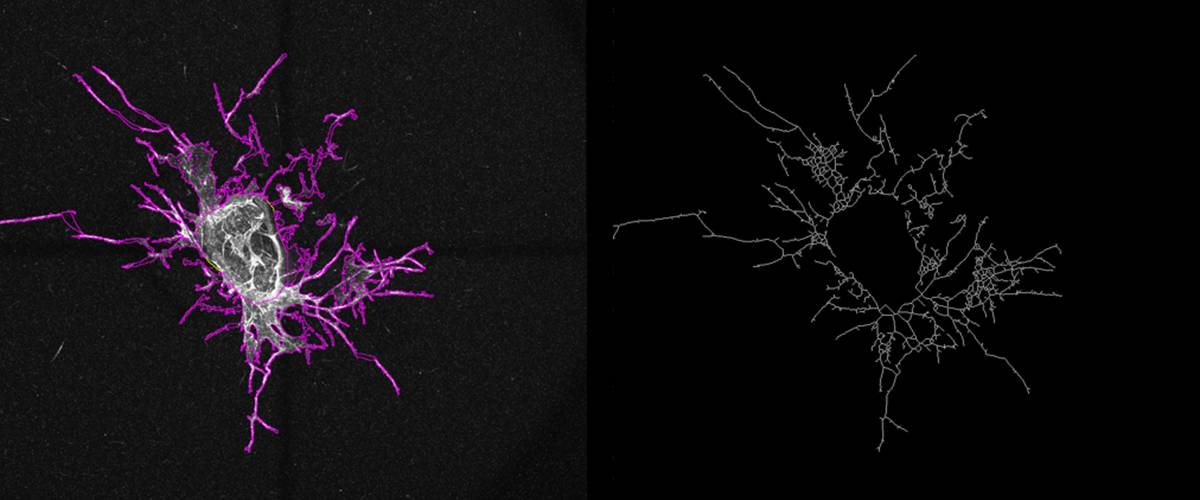
Quantification of angiogenic vessel growth from a segment of the aorta.
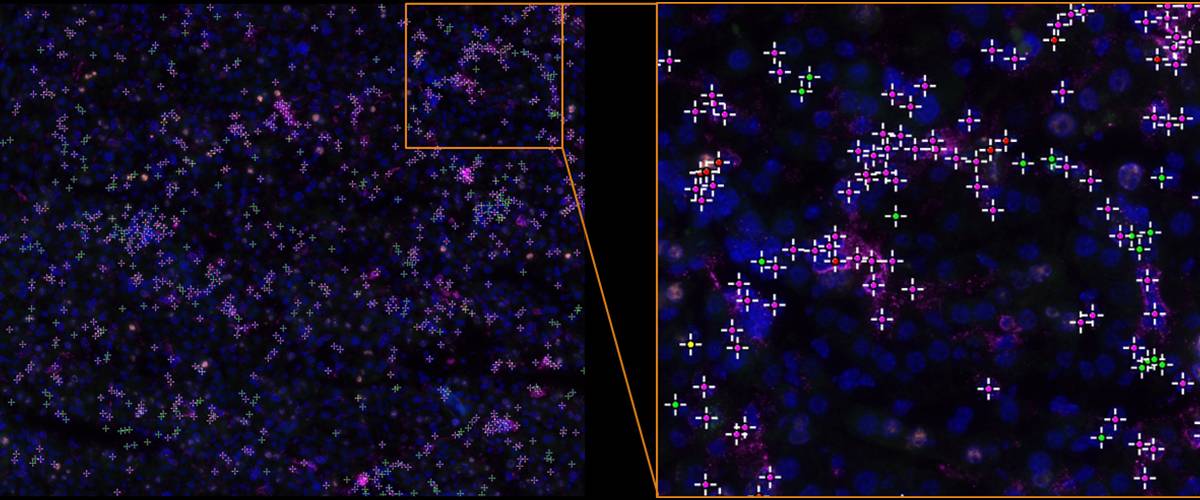
Quantification of cell populations in multiplex fluorescence imaging.
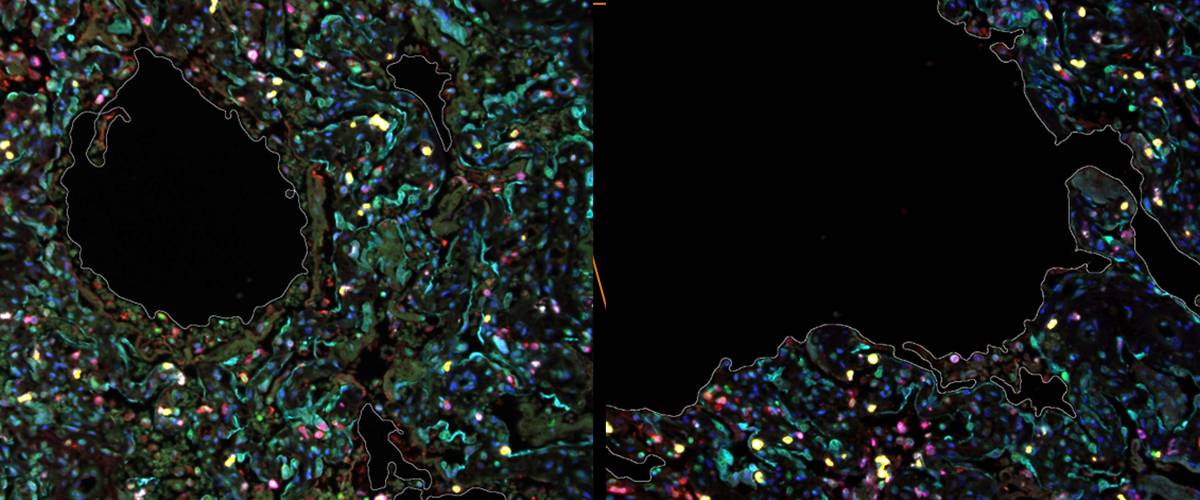
Detection of NETs in multiplex fluorescence imaging by marker localization.
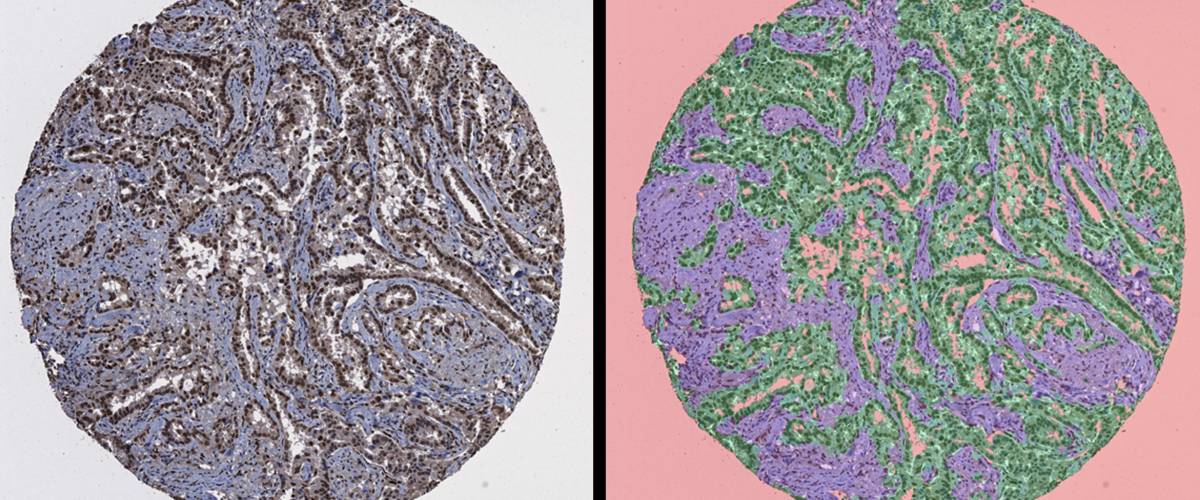
Automatic segmentation of different regions of tumor tissue using machine learning techniques.
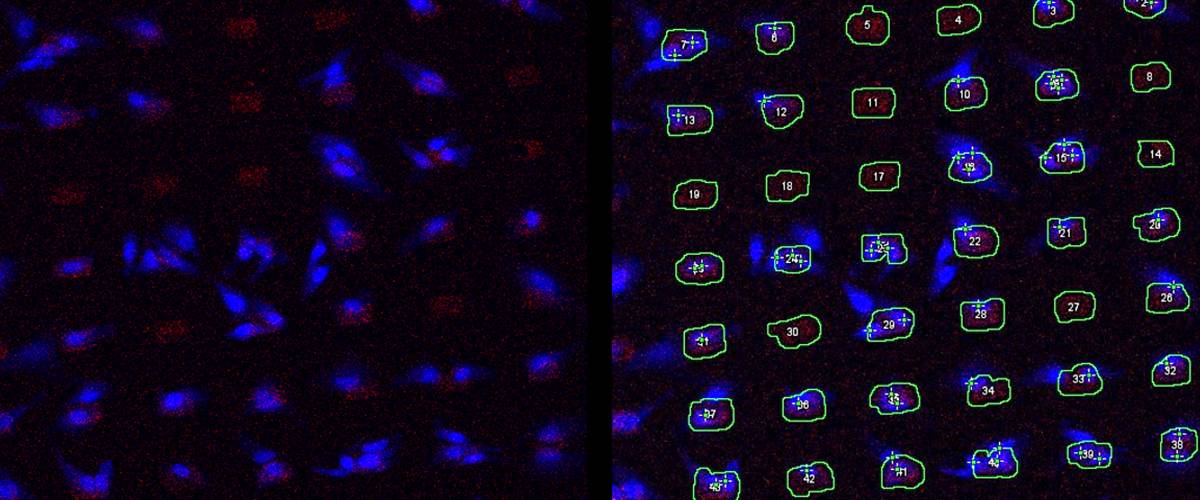
Detection of tumor cells on a deposited pattern by microcontact printing
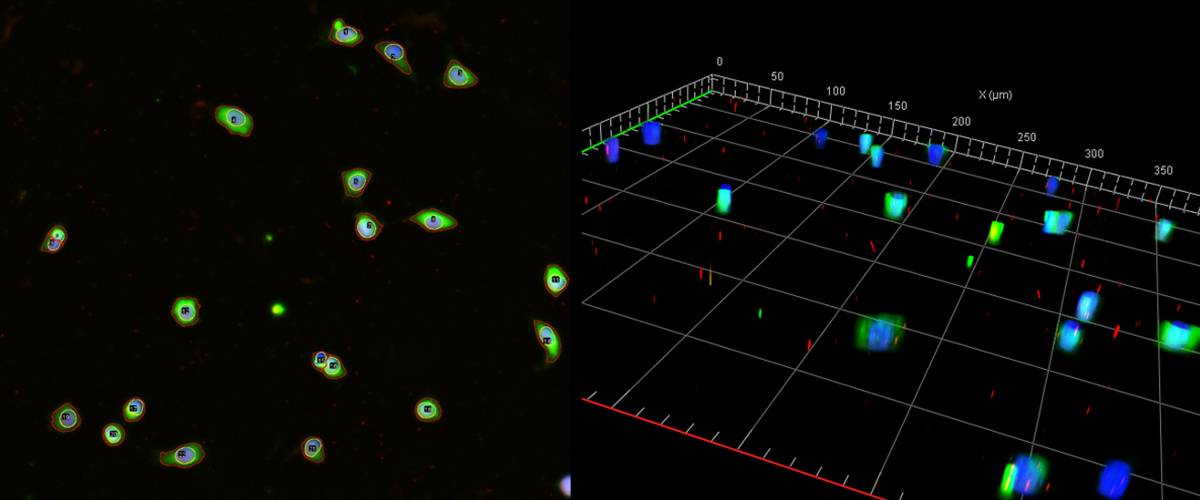
3D quantification of protein expression in nucleus and cytoplasm in cell cultures.
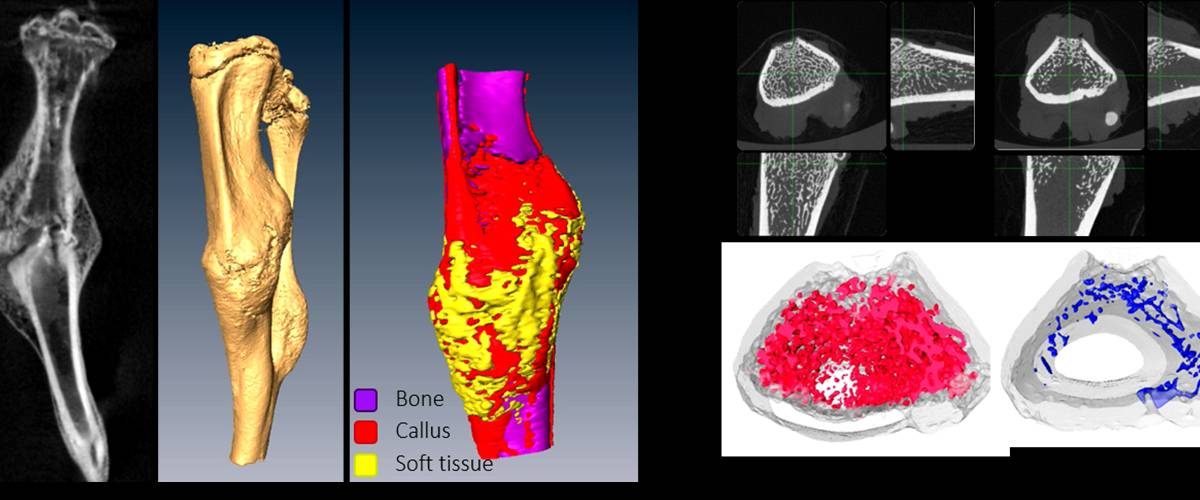
3D segmentation and quantification of bone structures in CT images. Quantification of bone volume and density, and histomorphological parameters in the case of trabecular bone structures.

3D segmentation and quantification of lungs and airways in CT images. Quantification of mean lung volume and intensity, with characterization of the degree of emphysema or infection.
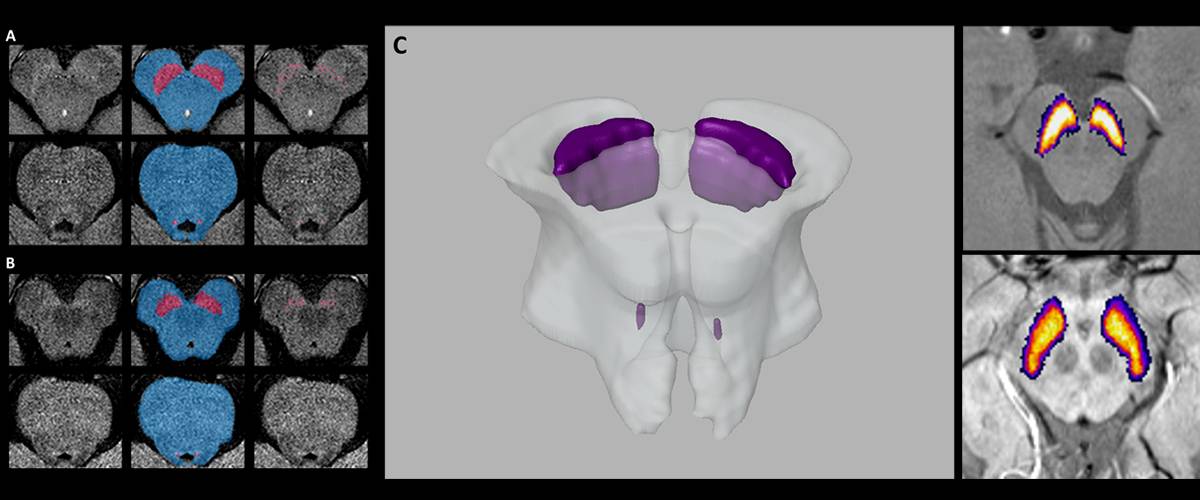
Segmentation and quantification of brain structures in MRI images.
Segmentation and 2D tracking of cells in microfluidic devices. Calculation of the track of each cell along the sequence and quantification of cell migration parameters.
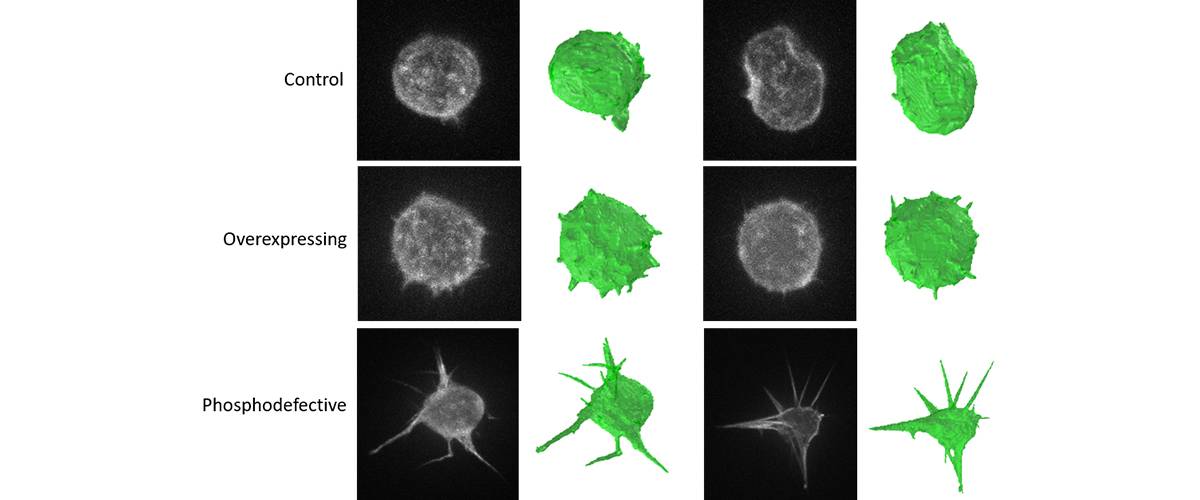
3D segmentation and characterization of cells by high resolution microscopy. Morphological quantification and detection of filopodia in different cell phenotypes.